
On the Bioanalyzer RNA-chips DNA will be visible in the size range from 4 to 10 kb. On an agarose gel, DNA contamination will be visible as a smear of band of fragments considerably larger than the RNA (>10 kb). Please make sure that your RNA isolation protocol employs a DNAse digestion step or other means to remove DNA from the sample. Provide at least 1 ug (2 ug preferred) of total RNA at a concentration of at least 50 ng/ul (1 ug for Poly-A enrichment 2 ug for ribo-depletion libraries using less starting material is possible, but we can’t guarantee results). Guidelines for Submission of Library-Worthy RNA Please see the Comprehensive Sample Requirements Page and consult our FAQs for technical questions
We do not perform RNA isolations for Illumina sequencing in our lab, but the PCR-lab in the neighboring building does. We require the submission of islotated total RNA samples.

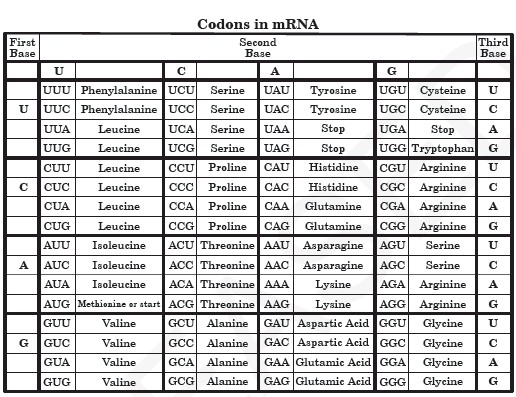
Please find information on single-cell RNA-seq here. The term “RNA-seq” is used for protocols that sequence the entire transcripts in contrast 3′-Tag-Seq sequences only one piece at the 3′-end. All our library preps are strand-specific. We offer RNA-seq library preparation, with multiple options such as ribo-depletion, poly-A enrichment, 3′-Tag-Seq (QuantSeq) libraries as described below as well as micro-RNA (miRNA) and small RNA library preps. Something of a misnomer because all the libraries end up as DNA, but this refers to the starting material.


 0 kommentar(er)
0 kommentar(er)
